SPARQLing-genomics
Today I would like to share the work I've been doing on SPARQLing-genomics -- an attempt to open the door to logic programming using RDF and SPARQL to data scientists.
Setting the stage
Its primary users are (data) scientists working primarily with DNA and RNA data. Unfortunately, most users are familiar with programming in R and/or Python, and fortunately GNU/Linux command-line tools.
A typical bioinformatics project in our group starts with sequencing data, in the form of FASTQ files. DNA data is then mapped to a reference genome and variants are called using a “standard” software pipeline, resulting in Variant Call Format (VCF) files.
At that point, analysis becomes non-standard, as the experiments differ in purpose and setup. This is the point where I think the RDF's simplicity can be of significant benefit to the data scientist, and those who attempt to understand the computational ground on which claims are made.
A tour of SPARQLing-genomics
The first thing to establish is information-parity between the “standard” pipeline output and its RDF equivalent. Naturally, developing tools to do that was my first priority.
Data preparation
Most data that needs further analysis exists in (multi-dimensional) tabular
format. The VCF is one of those multi-dimensional tabular formats. In
SPARQLing-genomics, a command-line tool called vcf2rdf can be used to
extract RDF triplets from this somewhat special format:
$ vcf2rdf -i variant_calls.vcf -O ntriples | gzip --stdout > variant_calls.n3.gzThis program lets the VCF file specify its own ontology, deferring the complexity of loose definitions to a time where we have SPARQL to connect the comparable attributes of variant calls.
Up to three-dimensional tabular data can be extracted using table2rdf:
$ table2rdf -i gene_regions.bed -O ntriples | gzip --stdout > gene_regions.n3.gzBoth vcfrdf and table2rdf use the SHA256 hash of the file's contents to
identify a file. This way, importing duplicate files won't lead to duplicate data.
For the large-scale people used to declarative programming, a convenience wrapper for folders exists:
$ folder2rdf --recursively --compress -i my_project/ -o my_rdf/ --threads 8This program attempts to find any data that can be imported and uses the appropriate tool to place the RDF equivalent of an input file in the output folder.
Once the RDF equivalents are available, they can be imported into a triple store. The instructions for this depend on the triple store.
Prototyping interface
I quickly realized that the lack of prototyping tools were a real time-waster for most users. When programming in R, there's RStudio, but when composing SPARQL queries, there's only a plain-text string or form field available to the users. That needed to change.
The prototyping interface needed to be easily accessible, multi-user, and get out of the way of a user's productivity. Therefore, I chose to implement it as a web service, meaning users need not to install or configure anything.
Having had positive experiences with Guile Scheme's web modules, I decided to write the web service in Guile Scheme.
To provide users with the ability to log in, without needing to create another account, I integrated it with our department's LDAP. Which brings us to the first page:
After logging in we're taken to an overview page:

This page is missing some flashy plots and interactive graphs. We need to save something for version 1.0.0, don't we?
In the spirit of Free Software, each statement has a “show me” button next to it, that leads to the query page with the query used to back up the statement:
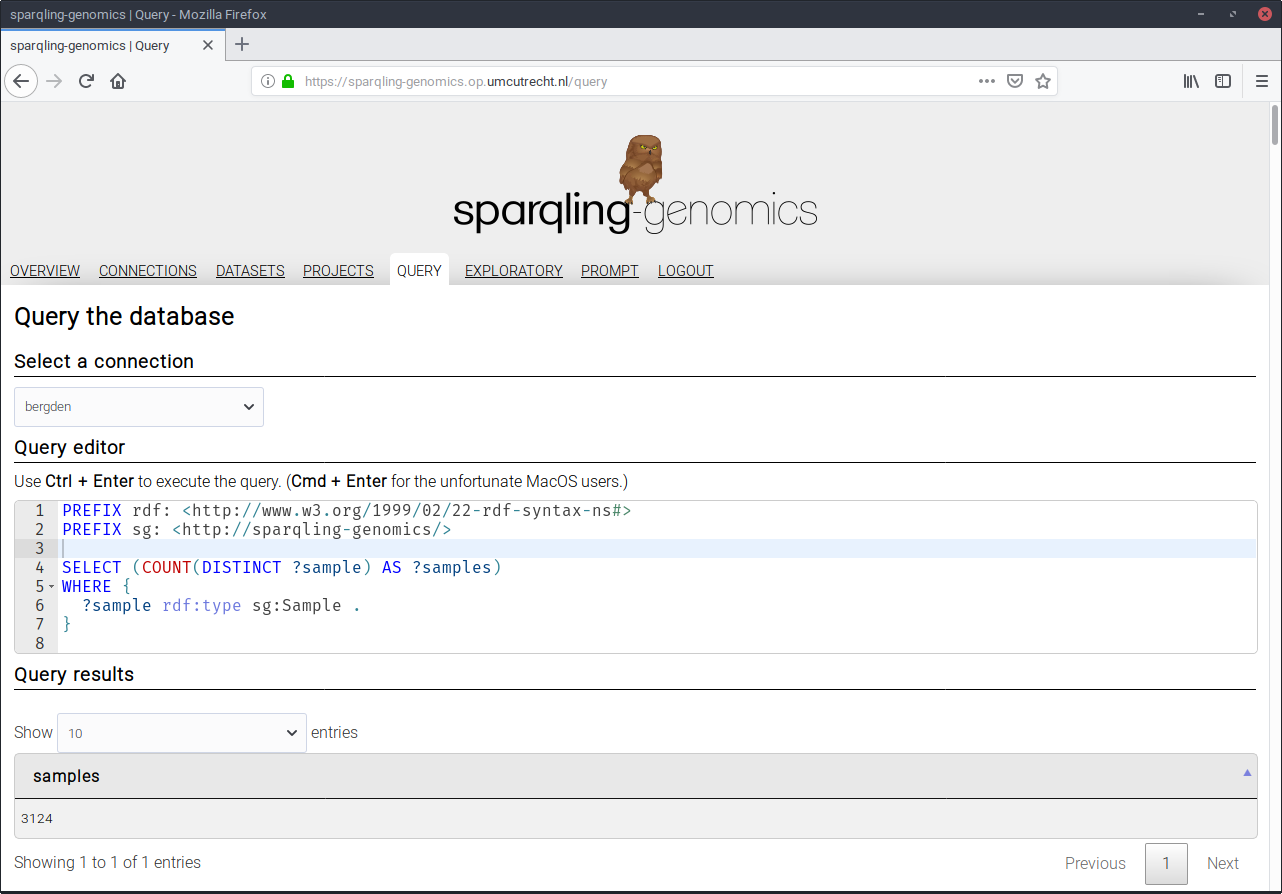
In this case, that there are 3124 samples available in the triple store. In future releases it would be interesting make plots that link back to their originating query, making the layer of software behind a plot more accessible with a single touch or click. There is still a lot to be gained in this area.
Talking about interesting features.. A side-effect of early user-testing is the development of obviously useful features like a query history of all successful queries:
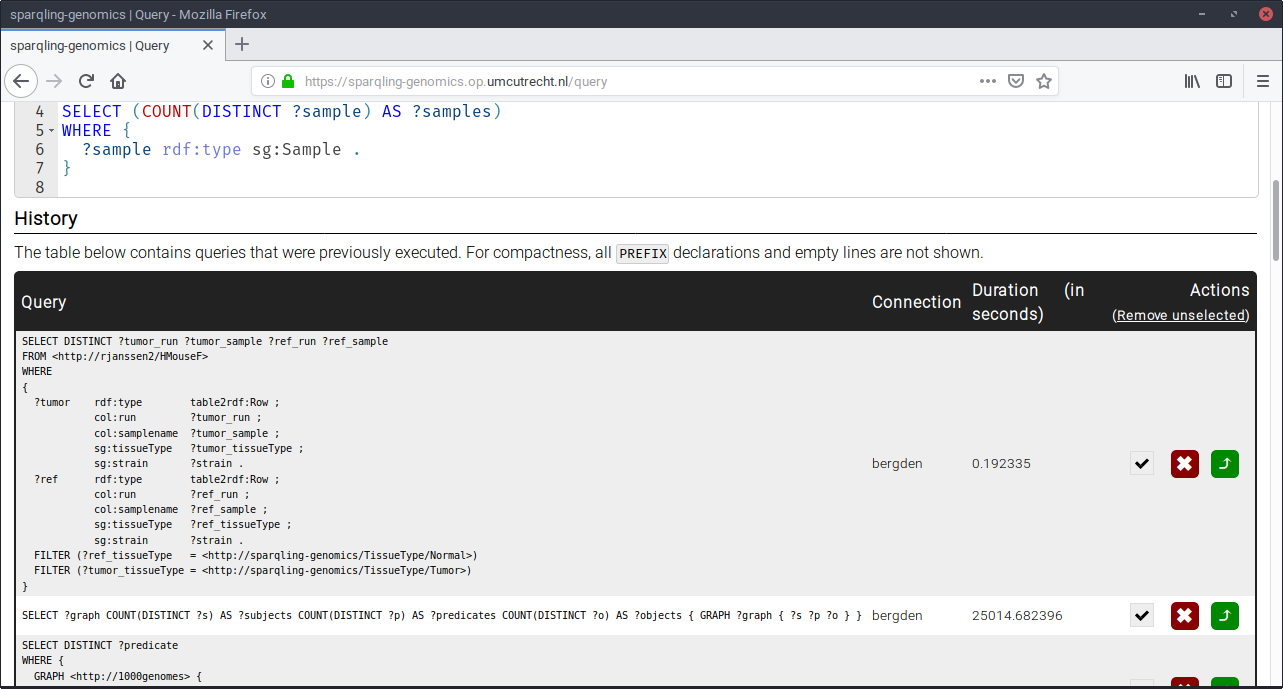
Before 1.0 we may want to add a “share with other user” button to enhance the experience for pair programming sessions.
Pair programming led to the development of something called the “exploratory”. To build a query pattern that matches the actual research question, two steps are often repeated:
- Discover the structure of how the triplets are connected to each other;
- Determine the relevant predicates for our real question.
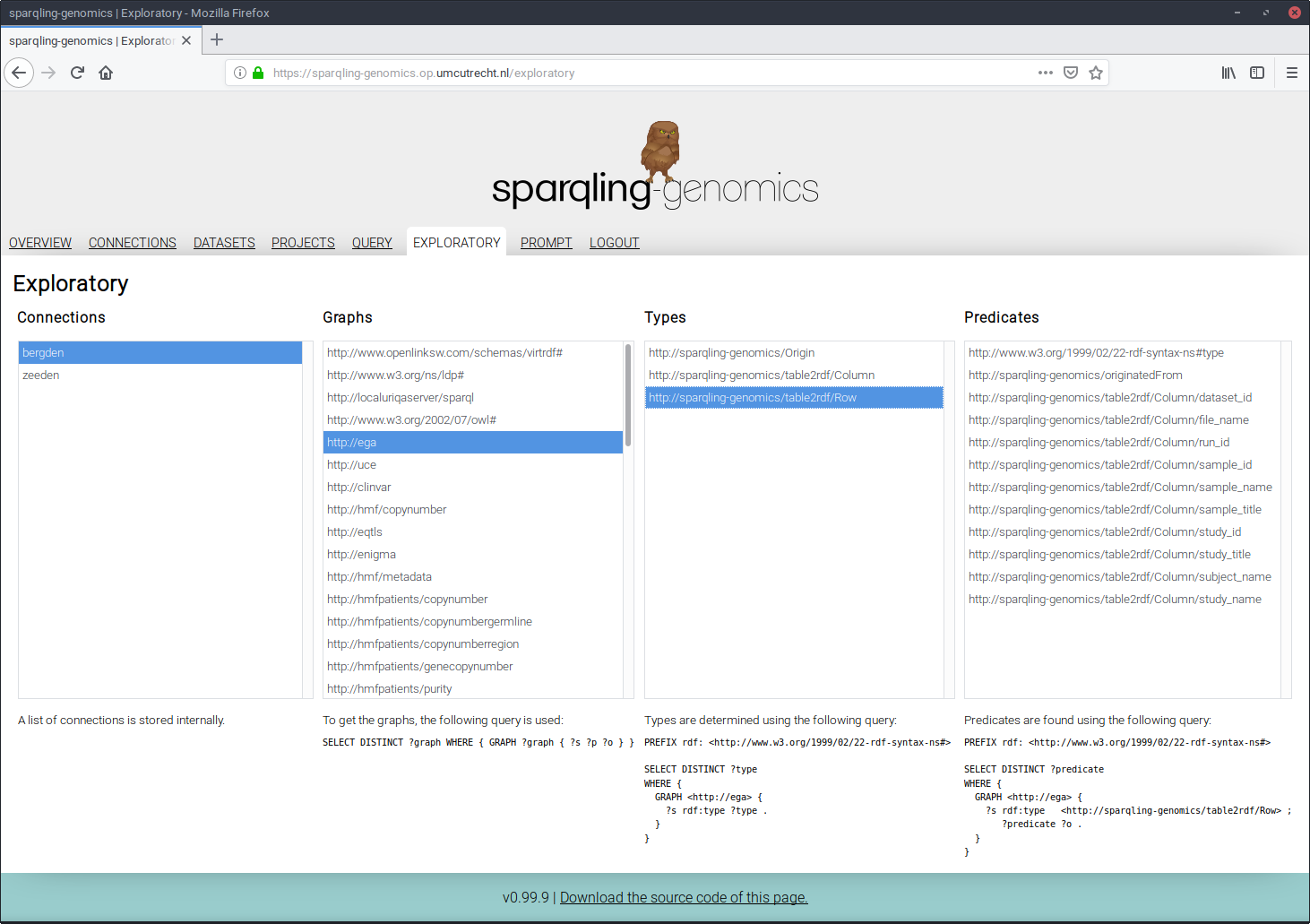
This connection -> graph -> types -> predicates structure makes finding predicates to use in reach within a few clicks rather than writing the queries. Note that the queries that lead to the list in each category is displayed below the category.
Prototyping only gets you so far
The web service provides various utilities to write the query that may lead to an answer to the original research question. Integrating it with other tools is a matter of copying the final query into the language of choice. I found that the integration into, for example, R, is quite seamless, because the result of a query is a table, which fits well in R's data frames. A user can use the SPARQL package in R, and execute the final query from the prototype, and move forward from there.
Wrapping up
I've tried to show you the highlights of SPARQLing-genomics. I spent a considerable amount of time writing documentation, which can be obtained from the Github page. It goes more in-depth on the command-line tools, and on how to deploy the web service on your own infrastructure.
